Gamma GEE
2017-05-04
Built using Zelig version 5.1.0.90000
Generalized Estimating Equation for Gamma Regression with gamma.gee.
The GEE gamma is similar to standard gamma regression (appropriate when you have an uncensored, positive-valued, continuous dependent variable such as the time until a parliamentary cabinet falls). Unlike in gamma regression, GEE gamma allows for dependence within clusters, such as in longitudinal data, although its use is not limited to just panel data. GEE models make no distributional assumptions but require three specifications: a mean function, a variance function, and a “working” correlation matrix for the clusters, which models the dependence of each observation with other observations in the same cluster. The “working” correlation matrix is a \(T \times T\) matrix of correlations, where \(T\) is the size of the largest cluster and the elements of the matrix are correlations between within-cluster observations. The appeal of GEE models is that it gives consistent estimates of the parameters and consistent estimates of the standard errors can be obtained using a robust “sandwich” estimator even if the “working” correlation matrix is incorrectly specified. If the “working” correlation matrix is correctly specified, GEE models will give more efficient estimates of the parameters. GEE models measure population-averaged effects as opposed to cluster-specific effects.
Syntax
With reference classes:
z5 <- zgammagee$new()
z5$zelig(Y ~ X1 + X2, model = "gamma.gee",
id = "X3", weights = w, data = mydata)
z5$setx()
z5$sim()With the Zelig 4 compatibility wrappers:
z.out <- zelig(Y ~ X1 + X2, model = "gamma.gee",
id = "X3", weights = w, data = mydata)
x.out <- setx(z.out)
s.out <- sim(z.out, x = x.out)where id is a variable which identifies the clusters. The data should be sorted by id and should be ordered within each cluster when appropriate.
Additional Inputs
Use the following arguments to specify the structure of the “working” correlations within clusters:
corstr: character string specifying the correlation structure: “independence”, “exchangeable”, “ar1”, “unstructured” and “userdefined”See
geeglmin packagegeepackfor other function arguments.
Examples
Example with Exchangeable Dependence
Attaching the sample turnout dataset:
data(coalition)Sorted variable identifying clusters
coalition$cluster <- c(rep(c(1:62), 5),rep(c(63), 4))
sorted.coalition <- coalition[order(coalition$cluster),]Estimating model and presenting summary:
z.out <- zelig(duration ~ fract + numst2, model = "gamma.gee",
id = "cluster", data = sorted.coalition,
corstr = "exchangeable")summary(z.out)## Model:
##
## Call:
## z5$zelig(formula = duration ~ fract + numst2, id = "cluster",
## corstr = "exchangeable", data = sorted.coalition)
##
## Coefficients:
## Estimate Std.err Wald Pr(>|W|)
## (Intercept) -1.296e-02 1.268e-02 1.045 0.3067
## fract 1.149e-04 1.474e-05 60.761 6.44e-15
## numst2 -1.740e-02 6.294e-03 7.643 0.0057
##
## Estimated Scale Parameters:
## Estimate Std.err
## (Intercept) 0.6231 0.04483
##
## Correlation: Structure = exchangeable Link = identity
##
## Estimated Correlation Parameters:
## Estimate Std.err
## alpha -0.008086 0.03363
## Number of clusters: 63 Maximum cluster size: 5
## Next step: Use 'setx' methodSetting the explanatory variables at their default values (mode for factor variables and mean for non-factor variables), with numst2 set to the vector 0 = no crisis, 1 = crisis.
Simulate quantities of interest
s.out <- sim(z.out, x = x.low, x1 = x.high)
summary(s.out)##
## sim x :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## [1,] 14.41 1.122 14.37 12.42 16.8
## pv
## mean sd 50% 2.5% 97.5%
## [1,] 14.56 17.89 8.322 0.068 65.74
##
## sim x1 :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## [1,] 19.2 1.067 19.16 17.22 21.39
## pv
## mean sd 50% 2.5% 97.5%
## [1,] 19.55 25.76 10.18 0.1106 92.71
## fd
## mean sd 50% 2.5% 97.5%
## [1,] 4.793 1.564 4.858 1.619 7.811Generate a plot of quantities of interest:
plot(s.out)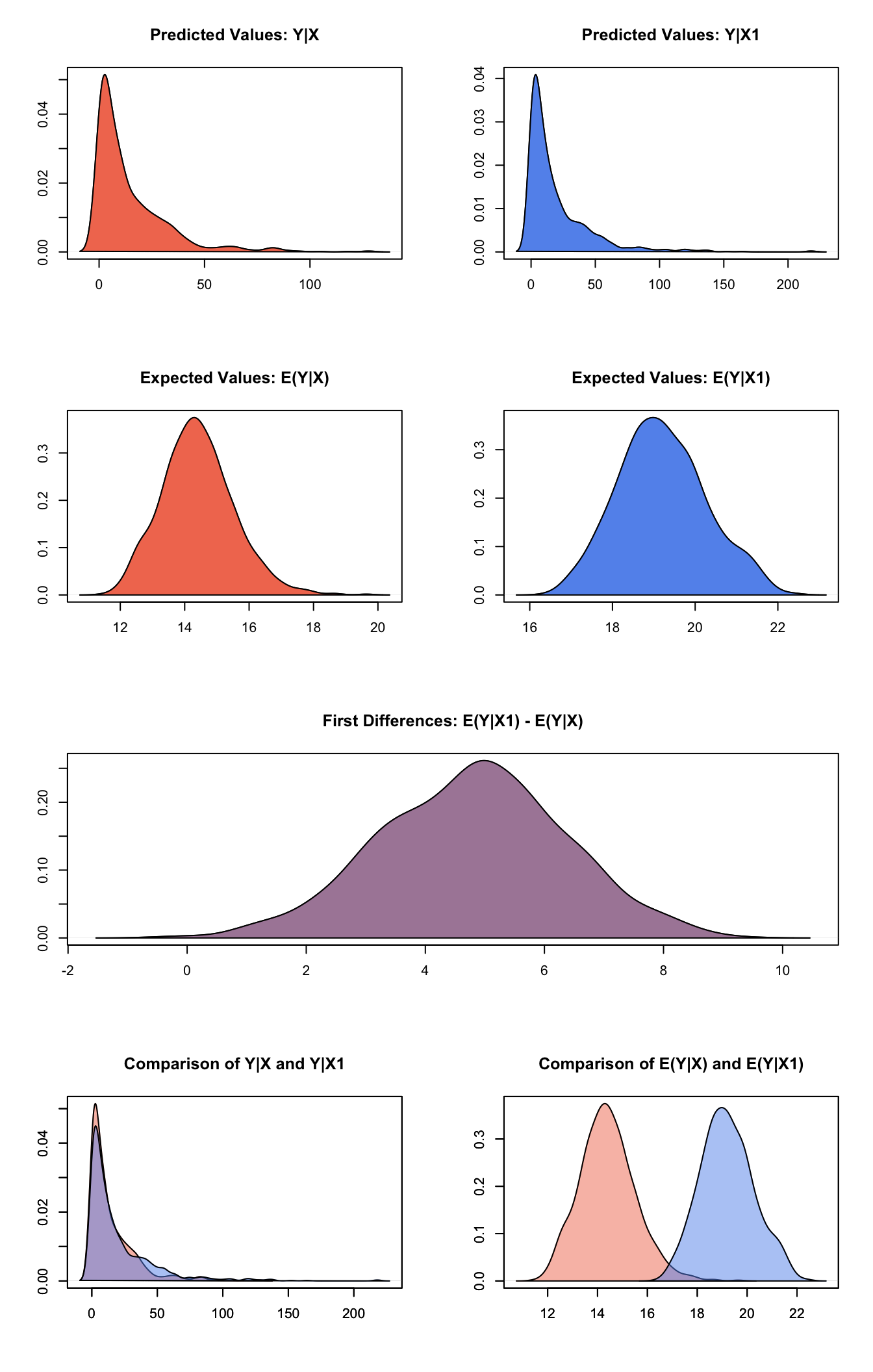
Graphs of Quantities of Interest for Gamma GEE
The Model
Suppose we have a panel dataset, with \(Y_{it}\) denoting the positive-valued, continuous dependent variable for unit \(i\) at time \(t\). \(Y_{i}\) is a vector or cluster of correlated data where \(y_{it}\) is correlated with \(y_{it^\prime}\) for some or all \(t, t^\prime\). Note that the model assumes correlations within \(i\) but independence across \(i\).
- The stochastic component is given by the joint and marginal distributions
where \(f\) and \(g\) are unspecified distributions with means \(\lambda_{i}\) and \(\lambda_{it}\). GEE models make no distributional assumptions and only require three specifications: a mean function, a variance function, and a correlation structure.
- The systematic component is the mean function, given by:
\[ \lambda_{it} = \frac{1}{x_{it} \beta} \]
where \(x_{it}\) is the vector of \(k\) explanatory variables for unit \(i\) at time \(t\) and \(\beta\) is the vector of coefficients.
- The variance function is given by:
\[ V_{it} = \lambda_{it}^2 = \frac{1}{(x_{it} \beta)^2} \]
- The correlation structure is defined by a \(T \times T\) “working” correlation matrix, where \(T\) is the size of the largest cluster. Users must specify the structure of the “working” correlation matrix a priori. The “working” correlation matrix then enters the variance term for each \(i\), given by:
\[ V_{i} = \phi \, A_{i}^{\frac{1}{2}} R_{i}(\alpha) A_{i}^{\frac{1}{2}} \]
where \(A_{i}\) is a \(T \times T\) diagonal matrix with the variance function \(V_{it} = \lambda_{it}^2\) as the \(t\) th diagonal element, \(R_{i}(\alpha)\) is the “working” correlation matrix, and \(\phi\) is a scale parameter. The parameters are then estimated via a quasi-likelihood approach.
- In GEE models, if the mean is correctly specified, but the variance and correlation structure are incorrectly specified, then GEE models provide consistent estimates of the parameters and thus the mean function as well, while consistent estimates of the standard errors can be obtained via a robust “sandwich” estimator. Similarly, if the mean and variance are correctly specified but the correlation structure is incorrectly specified, the parameters can be estimated consistently and the standard errors can be estimated consistently with the sandwich estimator. If all three are specified correctly, then the estimates of the parameters are more efficient.
Quantities of Interest
All quantities of interest are for marginal means rather than joint means.
The method of bootstrapping generally should not be used in GEE models. If you must bootstrap, bootstrapping should be done within clusters, which is not currently supported in Zelig. For conditional prediction models, data should be matched within clusters.
The expected values (qi$ev) for the GEE gamma model is the mean:
\[ E(Y) = \lambda_{c} = \frac{1}{x_c \beta}, \]
given draws of \(\beta\) from its sampling distribution, where \(x_{c}\) is a vector of values, one for each independent variable, chosen by the user.
- The first difference (qi$fd) for the GEE gamma model is defined as
\[ \textrm{FD} = \Pr(Y = 1 \mid x_1) - \Pr(Y = 1 \mid x). \]
- In conditional prediction models, the average expected treatment effect (att.ev) for the treatment group is
\[ \frac{1}{\sum_{i=1}^n \sum_{t=1}^T tr_{it}}\sum_{i:tr_{it}=1}^n \sum_{t:tr_{it}=1}^T \left\{ Y_{it}(tr_{it}=1) - E[Y_{it}(tr_{it}=0)] \right\}, \]
where \(tr_{it}\) is a binary explanatory variable defining the treatment (\(tr_{it}=1\)) and control (\(tr_{it}=0\)) groups. Variation in the simulations are due to uncertainty in simulating \(E[Y_{it}(tr_{it}=0)]\), the counterfactual expected value of \(Y_{it}\) for observations in the treatment group, under the assumption that everything stays the same except that the treatment indicator is switched to \(tr_{it}=0\).
Output Values
The output of each Zelig command contains useful information which you may view. For example, if you run
z.out <- zelig(y ~ x, model = "gamma.gee", id, data)then you may see a default summary of information through summary(z.out). Other elements available through the $ operator are listed below.
From the zelig() output object z.out, you may extract:
coefficients: parameter estimates for the explanatory variables.
residuals: the working residuals in the final iteration of the fit.
fitted.values: the vector of fitted values for the systemic component.
linear.predictors: the vector of \(x_{it}\beta\)
max.id: the size of the largest cluster.
From summary(z.out), you may extract:
coefficients: the parameter estimates with their associated standard errors, \(p\)-values, and \(z\)-statistics.
working.correlation: the “working” correlation matrix
From the sim() output object s.out, you may extract quantities of interest arranged as matrices indexed by simulation \(\times\) x-observation (for more than one x-observation). Available quantities are:
- qi$ev: the simulated expected values for the specified values of
qi$fd: the simulated first difference in the expected probabilities for the values specified in x and x1.
qi$att.ev: the simulated average expected treatment effect for the treated from conditional prediction models.
See also
The geeglm function is part of the geepack package by Søren Højsgaard, Ulrich Halekoh and Jun Yan. Advanced users may wish to refer to help(geepack) and help(family).
Højsgaard S, Halekoh U and Yan J (2006). “The R Package geepack for Generalized Estimating Equations.” Journal of Statistical Software, 15/2, pp. 1-11.
Yan J and Fine JP (2004). “Estimating Equations for Association Structures.” Statistics in Medicine, 23, pp. 859-880.
Yan J (2002). “geepack: Yet Another Package for Generalized Estimating Equations.” R-News, 2/3, pp. 12-14.