Probit Regression for Dependent Variables with Survey Weights
2017-05-04
Built using Zelig version 5.1.0.90000
Probit Regression for Dichotomous Dependent Variables with Survey Weights with probit.survey.
Use probit regression to model binary dependent variables specified as a function of a set of explanatory variables.
Examples
Example 1: User has Existing Sample Weights
Our example dataset comes from the survey package:
data(api, package = "survey")In this example, we will estimate a model using the percentages of students who receive subsidized lunch and the percentage who are new to a school to predict whether each California public school attends classes year round. We first make a numeric version of the variable in the example dataset, which you may not need to do in another dataset.
apistrat$yr.rnd.numeric <- as.numeric(apistrat$yr.rnd=="Yes")
z.out1 <- zelig(yr.rnd.numeric ~ meals + mobility,
model = "probit.survey", weights = apistrat$pw,
data = apistrat)## Warning in eval(family$initialize): non-integer #successes in a binomial
## glm!summary(z.out1)## Model:
##
## Call:
## z5$zelig(formula = yr.rnd.numeric ~ meals + mobility, data = apistrat,
## weights = apistrat$pw)
##
## Survey design:
## survey::svydesign(data = data, ids = ids, probs = probs, strata = strata,
## fpc = fpc, nest = nest, check.strata = check.strata, weights = localWeights)
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.980423 0.525356 -5.673 4.95e-08
## meals 0.017937 0.006358 2.821 0.00528
## mobility 0.046318 0.015090 3.069 0.00245
##
## (Dispersion parameter for binomial family taken to be 37.05532)
##
## Number of Fisher Scoring iterations: 7
##
## Next step: Use 'setx' methodSet explanatory variables to their default (mean/mode) values, and set a high (80th percentile) and low (20th percentile) value for “meals,” the percentage of students who receive subsidized meals:
x.low <- setx(z.out1, meals = quantile(apistrat$meals, 0.2))
x.high <- setx(z.out1, meals = quantile(apistrat$meals, 0.8))Generate first differences for the effect of high versus low “meals” on the probability that a school will hold classes year round:
s.out1 <- sim(z.out1, x=x.low, x1=x.high)
summary(s.out1)##
## sim x :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## [1,] 0.03510943 0.02665325 0.02791981 0.005130145 0.1057188
## pv
## 0 1
## [1,] 0.964 0.036
##
## sim x1 :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## [1,] 0.1886298 0.04351948 0.1858431 0.1109458 0.2808305
## pv
## 0 1
## [1,] 0.831 0.169
## fd
## mean sd 50% 2.5% 97.5%
## [1,] 0.1535203 0.04761432 0.151906 0.06034836 0.2519095Generate a second set of fitted values and a plot:
plot(s.out1)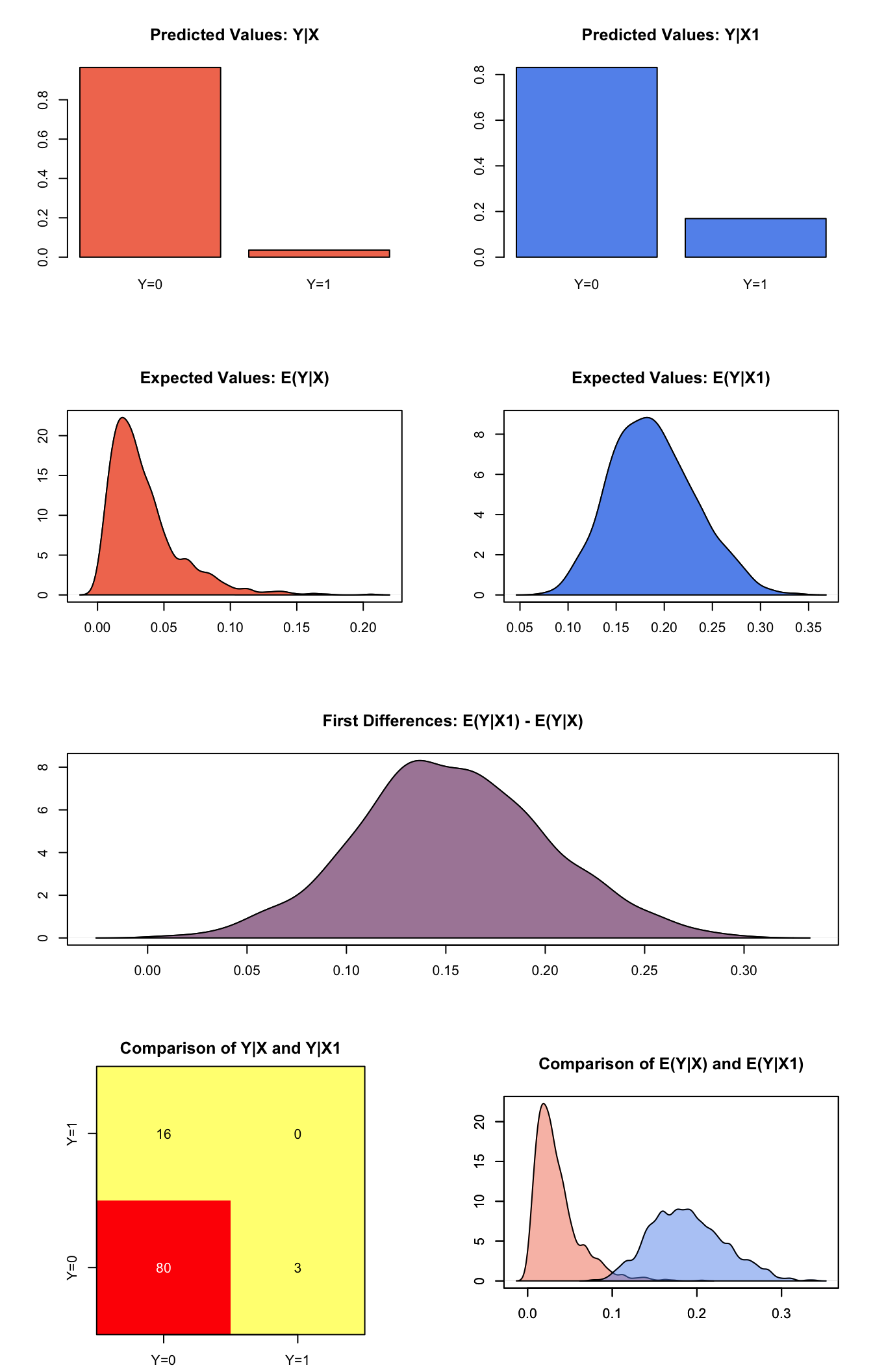
Graphs of Quantities of Interest for Probit Survey Model
Example 2: User has Details about Complex Survey Design (but not sample weights)
Suppose that the survey house that provided the dataset excluded probability weights but made other details about the survey design available. We can still estimate a model without probability weights that takes instead variables that identify each the stratum and/or cluster from which each observation was selected and the size of the finite sample from which each observation was selected.
z.out2 <- zelig(yr.rnd.numeric ~ meals + mobility, model = "probit.survey", strata=~stype, fpc=~fpc, data = apistrat)## Warning in eval(family$initialize): non-integer #successes in a binomial
## glm!summary(z.out2)## Model:
##
## Call:
## z5$zelig(formula = yr.rnd.numeric ~ meals + mobility, data = apistrat,
## strata = ~stype, fpc = ~fpc)
##
## Survey design:
## survey::svydesign(data = data, ids = ids, probs = probs, strata = strata,
## fpc = fpc, nest = nest, check.strata = check.strata, weights = localWeights)
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.886198 0.468368 -6.162 4.03e-09
## meals 0.019272 0.005933 3.249 0.001366
## mobility 0.033955 0.010109 3.359 0.000942
##
## (Dispersion parameter for binomial family taken to be 0.9377738)
##
## Number of Fisher Scoring iterations: 6
##
## Next step: Use 'setx' methodThe coefficient estimates from this model are identical to point estimates in the previous example, but the standard errors are smaller. When sampling weights are omitted, Zelig estimates them automatically for “normal.survey” models based on the user-defined description of sampling designs. In addition, when user-defined descriptions of the sampling design are entered as inputs, variance estimates are better and standard errors are consequently smaller.
The methods setx() and sim() can then be run on z.out2 in the same fashion described in Example 1.
Example 3: User has Replicate Weights
Load data for a model using the number of out-of-hospital cardiac arrests and the number of patients who arrive alive in hospitals to predict whether each hospital has been sued (an indicator variable artificially created here for the purpose of illustration).
data(scd, package = "survey")
scd$sued <- as.vector(c(0,0,0,1,1,1))Again, for the purpose of illustration, create four Balanced Repeated Replicate (BRR) weights:
BRRrep <- 2*cbind(c(1,0,1,0,1,0), c(1,0,0,1,0,1),
c(0,1,1,0,0,1), c(0,1,0,1,1,0))Estimate the model using Zelig:
z.out3 <- zelig(formula=sued ~ arrests + alive ,
model = "probit.survey",
repweights = BRRrep, type = "BRR", data = scd)## Warning in svrepdesign.default(data = data, repweights = repweights, type =
## type, : No sampling weights provided: equal probability assumed## Warning in eval(family$initialize): non-integer #successes in a binomial
## glm!
## Warning in eval(family$initialize): non-integer #successes in a binomial
## glm!## Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred## Warning in eval(family$initialize): non-integer #successes in a binomial
## glm!## Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred## Warning in eval(family$initialize): non-integer #successes in a binomial
## glm!## Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred## Warning in eval(family$initialize): non-integer #successes in a binomial
## glm!## Warning: glm.fit: fitted probabilities numerically 0 or 1 occurredsummary(z.out3)## Model:
##
## Call:
## z5$zelig(formula = sued ~ arrests + alive, data = scd, repweights = BRRrep,
## type = "BRR")
##
## Survey design:
## svrepdesign.default(data = data, repweights = repweights, type = type,
## weights = localWeights, combined.weights = combined.weights,
## rho = rho, bootstrap.average = bootstrap.average, scale = scale,
## rscales = rscales, fpctype = fpctype, fpc = fpc)
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.196e+01 7.874e-02 -278.85 0.00228
## arrests -5.907e-02 6.553e-04 -90.14 0.00706
## alive 8.509e-01 6.050e-03 140.64 0.00453
##
## (Dispersion parameter for binomial family taken to be 1.273459e-10)
##
## Number of Fisher Scoring iterations: 21
##
## Next step: Use 'setx' methodSet the explanatory variables at their means and set arrests at its 20th and 80th quartiles
x.low <- setx(z.out3, arrests = quantile(scd$arrests, .2))
x.high <- setx(z.out3, arrests = quantile(scd$arrests,.8))Generate first differences for the effect of the minimum versus the maximum number of individuals who arrive alive on the probability that a hospital will be sued:
s.out3 <- sim(z.out3, x = x.high, x1 = x.low)
summary(s.out3)##
## sim x :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## [1,] 2.220446e-16 0 2.220446e-16 2.220446e-16 2.220446e-16
## pv
## 0 1
## [1,] 1 0
##
## sim x1 :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## [1,] 1 0 1 1 1
## pv
## 0 1
## [1,] 0 1
## fd
## mean sd 50% 2.5% 97.5%
## [1,] 1 0 1 1 1Generate a second set of fitted values and a plot:
plot(s.out3)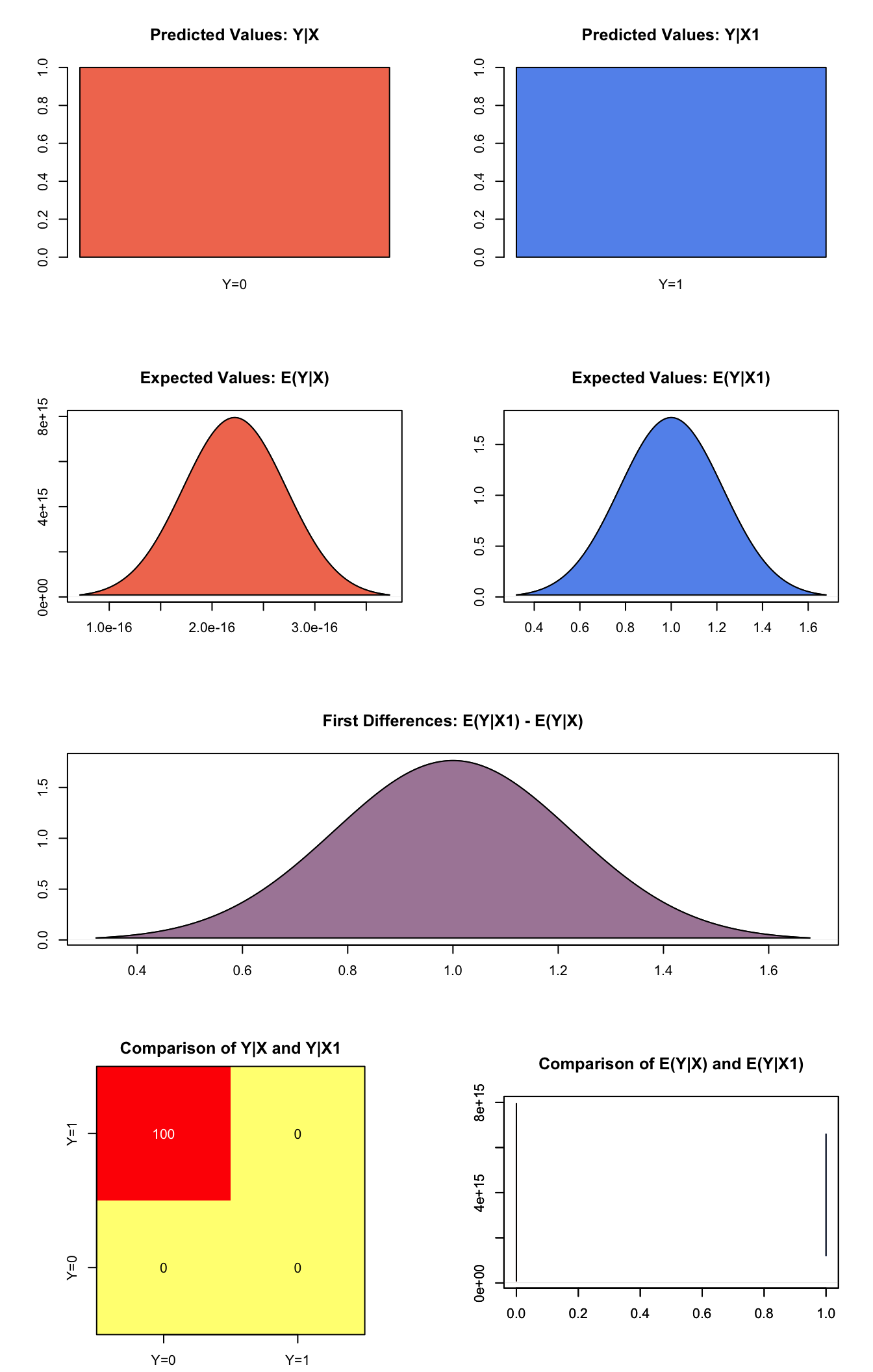
Graphs of Quantities of Interest for Probit Survey Model
The user should also refer to the probit model demo, since probit.survey models can take many of the same options as probit models.
Model
Let \(Y_i\) be the observed binary dependent variable for observation \(i\) which takes the value of either 0 or 1.
- The stochastic component is given by
\[ Y_i \; \sim \; \textrm{Bernoulli}(\pi_i), \]
where \(\pi_i=\Pr(Y_i=1)\).
- The systematic component is
\[ \pi_i \; = \; \Phi (x_i \beta) \]
where \(\Phi(\mu)\) is the cumulative distribution function of the Normal distribution with mean 0 and unit variance.
Quantities of Interest
- The expected value (qi$ev) is a simulation of predicted probability of success
\[ E(Y) = \pi_i = \Phi(x_i \beta), \]
given a draw of \(\beta\) from its sampling distribution.
The predicted value (qi$pr) is a draw from a Bernoulli distribution with mean \(\pi_i\).
The first difference (qi$fd) in expected values is defined as
\[ \textrm{FD} = \Pr(Y = 1 \mid x_1) - \Pr(Y = 1 \mid x). \]
- The risk ratio (qi$rr) is defined as
\[ \textrm{RR} = \Pr(Y = 1 \mid x_1) / \Pr(Y = 1 \mid x). \]
- In conditional prediction models, the average expected treatment effect (att.ev) for the treatment group is
\[ \frac{1}{\sum_{i=1}^n t_i}\sum_{i:t_i=1}^n \left\{ Y_i(t_i=1) - E[Y_i(t_i=0)] \right\}, \]
where \(t_i\) is a binary explanatory variable defining the treatment (\(t_i=1\)) and control (\(t_i=0\)) groups. Variation in the simulations are due to uncertainty in simulating \(E[Y_i(t_i=0)]\), the counterfactual expected value of \(Y_i\) for observations in the treatment group, under the assumption that everything stays the same except that the treatment indicator is switched to \(t_i=0\).
- In conditional prediction models, the average predicted treatment effect (att.pr) for the treatment group is
\[ \frac{1}{\sum_{i=1}^n t_i}\sum_{i:t_i=1}^n \left\{ Y_i(t_i=1) - \widehat{Y_i(t_i=0)} \right\}, \]
where \(t_i\) is a binary explanatory variable defining the treatment (\(t_i=1\)) and control (\(t_i=0\)) groups. Variation in the simulations are due to uncertainty in simulating \(\widehat{Y_i(t_i=0)}\), the counterfactual predicted value of \(Y_i\) for observations in the treatment group, under the assumption that everything stays the same except that the treatment indicator is switched to \(t_i=0\).
Output Values
The Zelig object stores fields containing everything needed to rerun the Zelig output, and all the results and simulations as they are generated. In addition to the summary commands demonstrated above, some simply utility functions (known as getters) provide easy access to the raw fields most commonly of use for further investigation.
In the example above z.out$get_coef() returns the estimated coefficients, z.out$get_vcov() returns the estimated covariance matrix, and z.out$get_predict() provides predicted values for all observations in the dataset from the analysis.
See also
The probitsurvey model is part of the survey package by Thomas Lumley, which in turn depends heavily on the glm package. Advanced users may wish to refer to help(glm) and help(family).
Lumley T (2016). “survey: analysis of complex survey samples.” R package version 3.31-5.
Lumley T (2004). “Analysis of Complex Survey Samples.” Journal of Statistical Software, 9 (1), pp. 1-19. R package verson 2.2.