Mutlinomial Logit
2017-05-04
Built using Zelig version 5.1.0.90000
Multinomial Logistic Regression for Dependent Variables with Unordered Categorical Values with mlogit in ZeligChoice.
Use the multinomial logit distribution to model unordered categorical variables. The dependent variable may be in the format of either character strings or integer values. See Multinomial Bayesian Logistic Regression for a Bayesian version of this model.
Syntax
First load packages:
library(zeligverse)With reference classes:
With the Zelig 4 compatibility wrappers:
z.out <- zelig(as.factor(Y) ~ X1 + X23,
model = "mlogit", data = mydata)
x.out <- setx(z.out)
s.out <- sim(z.out, x = x.out, x1 = NULL)where Y above is supposed to be a factor variable with levels apples,bananas,oranges. By default, oranges is the last level and omitted. (You cannot specify a different base level at this time.) For \(J\) equations, there must be \(J + 1\) levels.
Examples
Load the sample data:
data(mexico)Estimate the empirical model:
z.out1 <- zelig(as.factor(vote88) ~ pristr + othcok + othsocok,
model = "mlogit", data = mexico)Summarize estimated paramters:
summary(z.out1)## Model:
##
## Call:
## z5$zelig(formula = as.factor(vote88) ~ pristr + othcok + othsocok,
## data = mexico)
##
##
## Pearson residuals:
## Min 1Q Median 3Q Max
## log(mu[,1]/mu[,3]) -4.296 -0.6875 0.2791 0.7019 2.102
## log(mu[,2]/mu[,3]) -2.245 -0.4690 -0.2078 -0.0887 4.540
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept):1 2.87082 0.39638 7.243 4.40e-13
## (Intercept):2 0.39915 0.47013 0.849 0.3959
## pristr:1 0.59688 0.09124 6.542 6.07e-11
## pristr:2 -0.12503 0.10428 -1.199 0.2305
## othcok:1 -1.24259 0.11236 -11.059 < 2e-16
## othcok:2 -0.14066 0.13302 -1.057 0.2903
## othsocok:1 -0.30260 0.14960 -2.023 0.0431
## othsocok:2 0.04975 0.16097 0.309 0.7572
##
## Number of linear predictors: 2
##
## Names of linear predictors: log(mu[,1]/mu[,3]), log(mu[,2]/mu[,3])
##
## Residual deviance: 2360.571 on 2710 degrees of freedom
##
## Log-likelihood: -1180.286 on 2710 degrees of freedom
##
## Number of iterations: 4
##
## Reference group is level 3 of the response
## Next step: Use 'setx' methodSet the explanatory variables to their default values, with \(pristr\) (for the strength of the PRI) equal to 1 (weak) in the baseline values, and equal to 3 (strong) in the alternative values:
Generate simulated predicted probabilities qi$ev and differences in the predicted probabilities qi$fd:
s.out.mlogit <- sim(z.out1, x = x.strong, x1 = x.weak)
summary(s.out.mlogit)##
## sim x :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## Pr(Y=1) 0.7125607 0.02126999 0.7135892 0.6700842 0.7537237
## Pr(Y=2) 0.1288215 0.01480180 0.1277746 0.1026277 0.1603835
## Pr(Y=3) 0.1586178 0.01644167 0.1583075 0.1286804 0.1927543
## pv
## 1 2 3
## [1,] 0.7 0.137 0.163
##
## sim x1 :
## -----
## ev
## mean sd 50% 2.5% 97.5%
## Pr(Y=1) 0.4031843 0.02184860 0.4033209 0.3613922 0.4468739
## Pr(Y=2) 0.3040039 0.02187937 0.3037288 0.2645768 0.3488399
## Pr(Y=3) 0.2928118 0.02102680 0.2918071 0.2537728 0.3357307
## pv
## 1 2 3
## [1,] 0.408 0.283 0.309
## fd
## mean sd 50% 2.5% 97.5%
## Pr(Y=1) -0.3093764 0.03330788 -0.3098321 -0.37300500 -0.2447529
## Pr(Y=2) 0.1751824 0.02784848 0.1755189 0.12114315 0.2301688
## Pr(Y=3) 0.1341940 0.02792288 0.1345650 0.08144286 0.1892681plot(s.out.mlogit)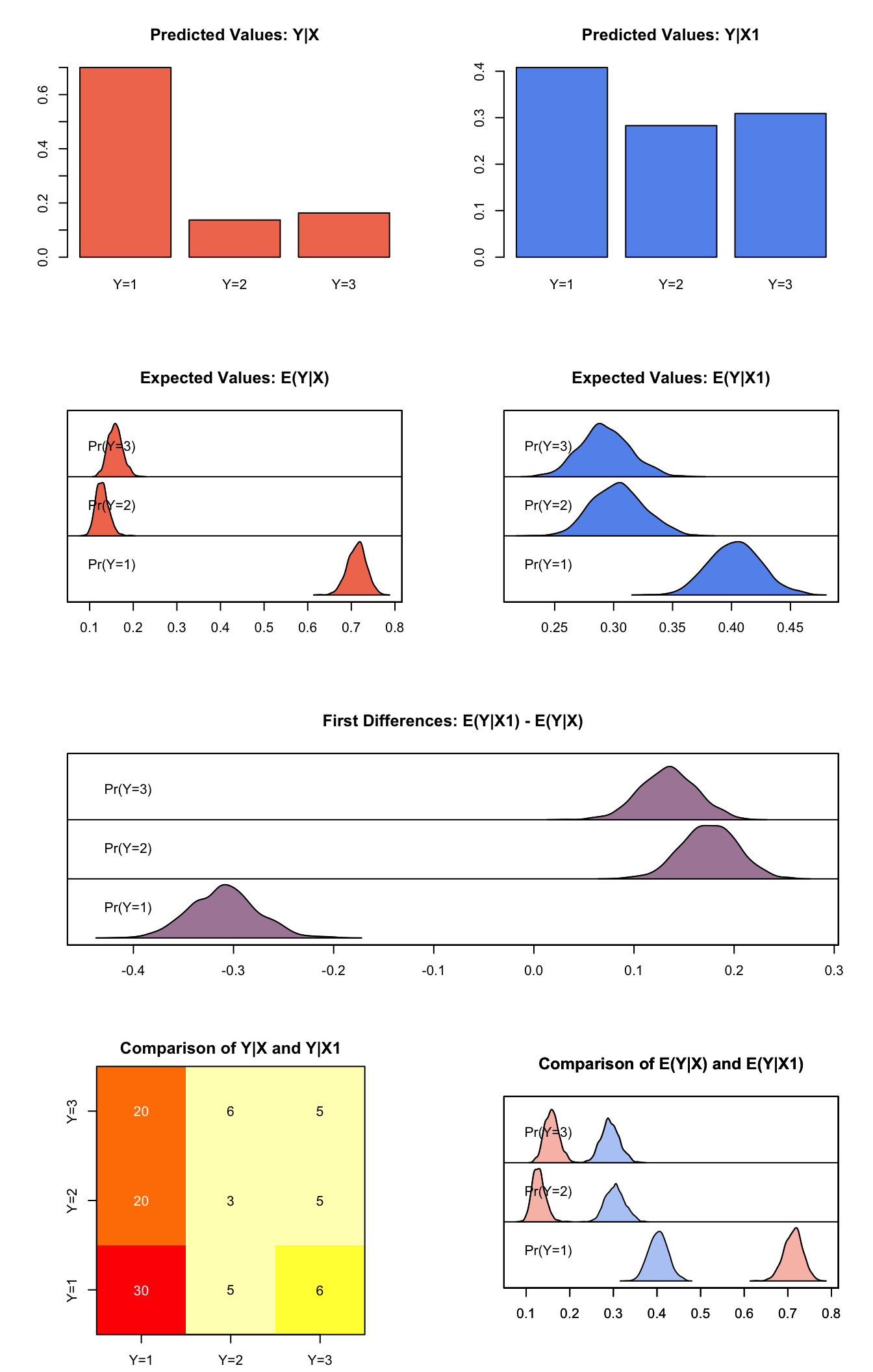
Graphs of Quantities of Interest for Multinomial Logit
Model
Let \(Y_i\) be the unordered categorical dependent variable that takes one of the values from 1 to \(J\), where \(J\) is the total number of categories.
- The stochastic component is given by
\[ Y_i \; \sim \; \textrm{Multinomial}(y_{i} \mid \pi_{ij}), \]
where \(\pi_{ij}=\Pr(Y_i=j)\) for \(j=1,\dots,J\).
- The systemic component is given by:
\[ \pi_{ij}\; = \; \frac{\exp(x_{i}\beta_{j})}{\sum^{J}_{k = 1} \exp(x_{i}\beta_{k})}, \]
where \(x_i\) is the vector of explanatory variables for observation \(i\), and \(\beta_j\) is the vector of coefficients for category \(j\).
Quantities of Interest
- The expected value (qi$ev) is the predicted probability for each category:
\[ E(Y) \; = \; \pi_{ij}\; = \; \frac{\exp(x_{i}\beta_{j})}{\sum^{J}_{k = 1} \exp(x_{i}\beta_{k})}. \]
The predicted value (qi$pr) is a draw from the multinomial distribution defined by the predicted probabilities.
The first difference in predicted probabilities (qi$fd), for each category is given by:
\[ \textrm{FD}_j = \Pr(Y=j \mid x_1) - \Pr(Y=j \mid x) \quad {\rm for} \quad j=1,\dots,J. \]
- In conditional prediction models, the average expected treatment effect (att.ev) for the treatment group is
\[ \frac{1}{n_j}\sum_{i:t_i=1}^{n_j} \left\{ Y_i(t_i=1) - E[Y_i(t_i=0)] \right\}, \]
where \(t_{i}\) is a binary explanatory variable defining the treatment (\(t_{i}=1\)) and control (\(t_{i}=0\)) groups, and \(n_j\) is the number of treated observations in category \(j\).
- In conditional prediction models, the average predicted treatment effect (att.pr) for the treatment group is
\[ \frac{1}{n_j}\sum_{i:t_i=1}^{n_j} \left\{ Y_i(t_i=1) - \widehat{Y_i(t_i=0)} \right\}, \]
where \(t_{i}\) is a binary explanatory variable defining the treatment (\(t_{i}=1\)) and control (\(t_{i}=0\)) groups, and \(n_j\) is the number of treated observations in category \(j\).
Output Values
The output of each Zelig command contains useful information which you may view. For example, if you run z.out <- zelig(y ~ x, model = mlogit, data), then you may examine the available information in z.out by using names(z.out), see the coefficients by using z.out\(coefficients, and a default summary of information through `summary(z.out)`. Other elements available through the `\)` operator are listed below.
From the `zelig()`` output object z.out, you may extract:
coefficients: the named vector of coefficients.
fitted.values: an \(n \times J\) matrix of the in-sample fitted values.
predictors: an \(n \times (J-1)\) matrix of the linear predictors \(x_i \beta_j\).
residuals: an \(n \times (J-1)\) matrix of the residuals.
df.residual: the residual degrees of freedom.
df.total: the total degrees of freedom.
rss: the residual sum of squares.
y: an \(n \times J\) matrix of the dependent variables.
zelig.data: the input data frame if save.data = TRUE.
From summary(z.out), you may extract:
coef3: a table of the coefficients with their associated standard errors and \(t\)-statistics.
cov.unscaled: the variance-covariance matrix.
pearson.resid: an \(n \times (m-1)\) matrix of the Pearson residuals.
See also
The multinomial logit function is part of the VGAM package by Thomas Yee. In addition, advanced users may wish to refer to help(vglm) in the VGAM library.
Yee TW (2015). Vector Generalized Linear and Additive Models: With an Implementation in R. Springer, New York, USA.
Yee TW and Wild CJ (1996). “Vector Generalized Additive Models.” Journal of Royal Statistical Society, Series B, 58 (3), pp. 481-493.
Yee TW (2010). “The VGAM Package for Categorical Data Analysis.” Journal of Statistical Software, 32 (10), pp. 1-34. <URL: http://www.jstatsoft.org/v32/i10/>.
Yee TW and Hadi AF (2014). “Row-column interaction models, with an R implementation.” Computational Statistics, 29 (6), pp. 1427-1445.
Yee TW (2017). VGAM: Vector Generalized Linear and Additive Models. R package version 1.0-3, <URL: https://CRAN.R-project.org/package=VGAM>.
Yee TW (2013). “Two-parameter reduced-rank vector generalized linear models.” Computational Statistics and Data Analysis. <URL: http://ees.elsevier.com/csda>.
Yee TW, Stoklosa J and Huggins RM (2015). “The VGAM Package for Capture-Recapture Data Using the Conditional Likelihood.” Journal of Statistical Software, 65 (5), pp. 1-33. <URL: http://www.jstatsoft.org/v65/i05/>.